# load the relevant tidymodels libraries
library(tidymodels)
library(tidyverse)
library(workflows)
library(tune)There’s a new modeling pipeline in town: tidymodels. Over the past few years, tidymodels has been gradually emerging as the tidyverse’s machine learning toolkit.
Why tidymodels? Well, it turns out that R has a consistency problem. Since everything was made by different people and using different principles, everything has a slightly different interface, and trying to keep everything in line can be frustrating. Several years ago, Max Kuhn (formerly at Pfeizer, now at RStudio) developed the caret R package (see my caret tutorial) aimed at creating a uniform interface for the massive variety of machine learning models that exist in R. Caret was great in a lot of ways, but also limited in others. In my own use, I found it to be quite slow whenever I tried to use on problems of any kind of modest size.
That said, caret was a great starting point, so RStudio hired Max Kuhn to work on a tidy version of caret, and he and many other people have developed what has become tidymodels. Tidymodels has been in development for a few years, with snippets of it being released as they were developed (see my post on the recipes package). I’ve been holding off writing a post about tidymodels until it seemed as though the different pieces fit together sufficiently for it to all feel cohesive. I feel like they’re finally there - which means it is time for me to learn it! While caret isn’t going anywhere (you can continue to use caret, and your existing caret code isn’t going to stop working), tidymodels will eventually make it redundant.
The main resources I used to learn tidymodels were Alison Hill’s slides from Introduction to Machine Learning with the Tidyverse, which contains all the slides for the course she prepared with Garrett Grolemund for RStudio::conf(2020), and Edgar Ruiz’s Gentle introduction to tidymodels on the RStudio website.
Note that throughout this post I’ll be assuming basic tidyverse knowledge, primarily of dplyr (e.g. piping %>% and function such as mutate()). Fortunately, for all you purrr-phobes out there, purrr is not required. If you’d like to brush up on your tidyverse skills, check out my Introduction to the Tidyverse posts. If you’d like to learn purrr (purrr is very handy for working with tidymodels but is no longer a requirement), check out my purrr post.
What is tidymodels
Much like the tidyverse consists of many core packages, such as ggplot2 and dplyr, tidymodels also consists of several core packages, including
rsample: for sample splitting (e.g. train/test or cross-validation)recipes: for pre-processingparsnip: for specifying the modelyardstick: for evaluating the model
Similarly to how you can load the entire tidyverse suite of packages by typing library(tidyverse), you can load the entire tidymodels suite of packages by typing library(tidymodels).
We will also be using the tune package (for parameter tuning procedure) and the workflows package (for putting everything together) that I had thought were a part of CRAN’s tidymodels package bundle, but apparently they aren’t. These will need to be loaded separately for now.
Unlike in my tidyverse post, I won’t base this post around the packages themselves, but I will mention the packages in passing.
Getting set up
First we need to load some libraries: tidymodels and tidyverse.
If you don’t already have the tidymodels library (or any of the other libraries) installed, then you’ll need to install it (once only) using install.packages("tidymodels").
We will use the Pima Indian Women’s diabetes dataset which contains information on 768 Pima Indian women’s diabetes status, as well as many predictive features such as the number of pregnancies (pregnant), plasma glucose concentration (glucose), diastolic blood pressure (pressure), triceps skin fold thickness (triceps), 2-hour serum insulin (insulin), BMI (mass), diabetes pedigree function (pedigree), and their age (age). In case you were wondering, the Pima Indians are a group of Native Americans living in an area consisting of what is now central and southern Arizona. The short name, “Pima” is believed to have come from a phrase meaning “I don’t know,” which they used repeatedly in their initial meetings with Spanish colonists. Thanks Wikipedia!
# load the Pima Indians dataset from the mlbench dataset
library(mlbench)
data(PimaIndiansDiabetes)
# rename dataset to have shorter name because lazy
diabetes_orig <- PimaIndiansDiabetesdiabetes_orig pregnant glucose pressure triceps insulin mass pedigree age diabetes
1 6 148 72 35 0 33.6 0.627 50 pos
2 1 85 66 29 0 26.6 0.351 31 neg
3 8 183 64 0 0 23.3 0.672 32 pos
4 1 89 66 23 94 28.1 0.167 21 neg
5 0 137 40 35 168 43.1 2.288 33 pos
6 5 116 74 0 0 25.6 0.201 30 neg
7 3 78 50 32 88 31.0 0.248 26 pos
8 10 115 0 0 0 35.3 0.134 29 neg
9 2 197 70 45 543 30.5 0.158 53 pos
10 8 125 96 0 0 0.0 0.232 54 pos
11 4 110 92 0 0 37.6 0.191 30 neg
12 10 168 74 0 0 38.0 0.537 34 pos
13 10 139 80 0 0 27.1 1.441 57 neg
14 1 189 60 23 846 30.1 0.398 59 pos
15 5 166 72 19 175 25.8 0.587 51 pos
16 7 100 0 0 0 30.0 0.484 32 pos
17 0 118 84 47 230 45.8 0.551 31 pos
18 7 107 74 0 0 29.6 0.254 31 pos
19 1 103 30 38 83 43.3 0.183 33 neg
20 1 115 70 30 96 34.6 0.529 32 pos
21 3 126 88 41 235 39.3 0.704 27 neg
22 8 99 84 0 0 35.4 0.388 50 neg
23 7 196 90 0 0 39.8 0.451 41 pos
24 9 119 80 35 0 29.0 0.263 29 pos
25 11 143 94 33 146 36.6 0.254 51 pos
26 10 125 70 26 115 31.1 0.205 41 pos
27 7 147 76 0 0 39.4 0.257 43 pos
28 1 97 66 15 140 23.2 0.487 22 neg
29 13 145 82 19 110 22.2 0.245 57 neg
30 5 117 92 0 0 34.1 0.337 38 neg
31 5 109 75 26 0 36.0 0.546 60 neg
32 3 158 76 36 245 31.6 0.851 28 pos
33 3 88 58 11 54 24.8 0.267 22 neg
34 6 92 92 0 0 19.9 0.188 28 neg
35 10 122 78 31 0 27.6 0.512 45 neg
36 4 103 60 33 192 24.0 0.966 33 neg
37 11 138 76 0 0 33.2 0.420 35 neg
38 9 102 76 37 0 32.9 0.665 46 pos
39 2 90 68 42 0 38.2 0.503 27 pos
40 4 111 72 47 207 37.1 1.390 56 pos
41 3 180 64 25 70 34.0 0.271 26 neg
42 7 133 84 0 0 40.2 0.696 37 neg
43 7 106 92 18 0 22.7 0.235 48 neg
44 9 171 110 24 240 45.4 0.721 54 pos
45 7 159 64 0 0 27.4 0.294 40 neg
46 0 180 66 39 0 42.0 1.893 25 pos
47 1 146 56 0 0 29.7 0.564 29 neg
48 2 71 70 27 0 28.0 0.586 22 neg
49 7 103 66 32 0 39.1 0.344 31 pos
50 7 105 0 0 0 0.0 0.305 24 neg
51 1 103 80 11 82 19.4 0.491 22 neg
52 1 101 50 15 36 24.2 0.526 26 neg
53 5 88 66 21 23 24.4 0.342 30 neg
54 8 176 90 34 300 33.7 0.467 58 pos
55 7 150 66 42 342 34.7 0.718 42 neg
56 1 73 50 10 0 23.0 0.248 21 neg
57 7 187 68 39 304 37.7 0.254 41 pos
58 0 100 88 60 110 46.8 0.962 31 neg
59 0 146 82 0 0 40.5 1.781 44 neg
60 0 105 64 41 142 41.5 0.173 22 neg
61 2 84 0 0 0 0.0 0.304 21 neg
62 8 133 72 0 0 32.9 0.270 39 pos
63 5 44 62 0 0 25.0 0.587 36 neg
64 2 141 58 34 128 25.4 0.699 24 neg
65 7 114 66 0 0 32.8 0.258 42 pos
66 5 99 74 27 0 29.0 0.203 32 neg
67 0 109 88 30 0 32.5 0.855 38 pos
68 2 109 92 0 0 42.7 0.845 54 neg
69 1 95 66 13 38 19.6 0.334 25 neg
70 4 146 85 27 100 28.9 0.189 27 neg
71 2 100 66 20 90 32.9 0.867 28 pos
72 5 139 64 35 140 28.6 0.411 26 neg
73 13 126 90 0 0 43.4 0.583 42 pos
74 4 129 86 20 270 35.1 0.231 23 neg
75 1 79 75 30 0 32.0 0.396 22 neg
76 1 0 48 20 0 24.7 0.140 22 neg
77 7 62 78 0 0 32.6 0.391 41 neg
78 5 95 72 33 0 37.7 0.370 27 neg
79 0 131 0 0 0 43.2 0.270 26 pos
80 2 112 66 22 0 25.0 0.307 24 neg
81 3 113 44 13 0 22.4 0.140 22 neg
82 2 74 0 0 0 0.0 0.102 22 neg
83 7 83 78 26 71 29.3 0.767 36 neg
84 0 101 65 28 0 24.6 0.237 22 neg
85 5 137 108 0 0 48.8 0.227 37 pos
86 2 110 74 29 125 32.4 0.698 27 neg
87 13 106 72 54 0 36.6 0.178 45 neg
88 2 100 68 25 71 38.5 0.324 26 neg
89 15 136 70 32 110 37.1 0.153 43 pos
90 1 107 68 19 0 26.5 0.165 24 neg
91 1 80 55 0 0 19.1 0.258 21 neg
92 4 123 80 15 176 32.0 0.443 34 neg
93 7 81 78 40 48 46.7 0.261 42 neg
94 4 134 72 0 0 23.8 0.277 60 pos
95 2 142 82 18 64 24.7 0.761 21 neg
96 6 144 72 27 228 33.9 0.255 40 neg
97 2 92 62 28 0 31.6 0.130 24 neg
98 1 71 48 18 76 20.4 0.323 22 neg
99 6 93 50 30 64 28.7 0.356 23 neg
100 1 122 90 51 220 49.7 0.325 31 pos
101 1 163 72 0 0 39.0 1.222 33 pos
102 1 151 60 0 0 26.1 0.179 22 neg
103 0 125 96 0 0 22.5 0.262 21 neg
104 1 81 72 18 40 26.6 0.283 24 neg
105 2 85 65 0 0 39.6 0.930 27 neg
106 1 126 56 29 152 28.7 0.801 21 neg
107 1 96 122 0 0 22.4 0.207 27 neg
108 4 144 58 28 140 29.5 0.287 37 neg
109 3 83 58 31 18 34.3 0.336 25 neg
110 0 95 85 25 36 37.4 0.247 24 pos
111 3 171 72 33 135 33.3 0.199 24 pos
112 8 155 62 26 495 34.0 0.543 46 pos
113 1 89 76 34 37 31.2 0.192 23 neg
114 4 76 62 0 0 34.0 0.391 25 neg
115 7 160 54 32 175 30.5 0.588 39 pos
116 4 146 92 0 0 31.2 0.539 61 pos
117 5 124 74 0 0 34.0 0.220 38 pos
118 5 78 48 0 0 33.7 0.654 25 neg
119 4 97 60 23 0 28.2 0.443 22 neg
120 4 99 76 15 51 23.2 0.223 21 neg
121 0 162 76 56 100 53.2 0.759 25 pos
122 6 111 64 39 0 34.2 0.260 24 neg
123 2 107 74 30 100 33.6 0.404 23 neg
124 5 132 80 0 0 26.8 0.186 69 neg
125 0 113 76 0 0 33.3 0.278 23 pos
126 1 88 30 42 99 55.0 0.496 26 pos
127 3 120 70 30 135 42.9 0.452 30 neg
128 1 118 58 36 94 33.3 0.261 23 neg
129 1 117 88 24 145 34.5 0.403 40 pos
130 0 105 84 0 0 27.9 0.741 62 pos
131 4 173 70 14 168 29.7 0.361 33 pos
132 9 122 56 0 0 33.3 1.114 33 pos
133 3 170 64 37 225 34.5 0.356 30 pos
134 8 84 74 31 0 38.3 0.457 39 neg
135 2 96 68 13 49 21.1 0.647 26 neg
136 2 125 60 20 140 33.8 0.088 31 neg
137 0 100 70 26 50 30.8 0.597 21 neg
138 0 93 60 25 92 28.7 0.532 22 neg
139 0 129 80 0 0 31.2 0.703 29 neg
140 5 105 72 29 325 36.9 0.159 28 neg
141 3 128 78 0 0 21.1 0.268 55 neg
142 5 106 82 30 0 39.5 0.286 38 neg
143 2 108 52 26 63 32.5 0.318 22 neg
144 10 108 66 0 0 32.4 0.272 42 pos
145 4 154 62 31 284 32.8 0.237 23 neg
146 0 102 75 23 0 0.0 0.572 21 neg
147 9 57 80 37 0 32.8 0.096 41 neg
148 2 106 64 35 119 30.5 1.400 34 neg
149 5 147 78 0 0 33.7 0.218 65 neg
150 2 90 70 17 0 27.3 0.085 22 neg
151 1 136 74 50 204 37.4 0.399 24 neg
152 4 114 65 0 0 21.9 0.432 37 neg
153 9 156 86 28 155 34.3 1.189 42 pos
154 1 153 82 42 485 40.6 0.687 23 neg
155 8 188 78 0 0 47.9 0.137 43 pos
156 7 152 88 44 0 50.0 0.337 36 pos
157 2 99 52 15 94 24.6 0.637 21 neg
158 1 109 56 21 135 25.2 0.833 23 neg
159 2 88 74 19 53 29.0 0.229 22 neg
160 17 163 72 41 114 40.9 0.817 47 pos
161 4 151 90 38 0 29.7 0.294 36 neg
162 7 102 74 40 105 37.2 0.204 45 neg
163 0 114 80 34 285 44.2 0.167 27 neg
164 2 100 64 23 0 29.7 0.368 21 neg
165 0 131 88 0 0 31.6 0.743 32 pos
166 6 104 74 18 156 29.9 0.722 41 pos
167 3 148 66 25 0 32.5 0.256 22 neg
168 4 120 68 0 0 29.6 0.709 34 neg
169 4 110 66 0 0 31.9 0.471 29 neg
170 3 111 90 12 78 28.4 0.495 29 neg
171 6 102 82 0 0 30.8 0.180 36 pos
172 6 134 70 23 130 35.4 0.542 29 pos
173 2 87 0 23 0 28.9 0.773 25 neg
174 1 79 60 42 48 43.5 0.678 23 neg
175 2 75 64 24 55 29.7 0.370 33 neg
176 8 179 72 42 130 32.7 0.719 36 pos
177 6 85 78 0 0 31.2 0.382 42 neg
178 0 129 110 46 130 67.1 0.319 26 pos
179 5 143 78 0 0 45.0 0.190 47 neg
180 5 130 82 0 0 39.1 0.956 37 pos
181 6 87 80 0 0 23.2 0.084 32 neg
182 0 119 64 18 92 34.9 0.725 23 neg
183 1 0 74 20 23 27.7 0.299 21 neg
184 5 73 60 0 0 26.8 0.268 27 neg
185 4 141 74 0 0 27.6 0.244 40 neg
186 7 194 68 28 0 35.9 0.745 41 pos
187 8 181 68 36 495 30.1 0.615 60 pos
188 1 128 98 41 58 32.0 1.321 33 pos
189 8 109 76 39 114 27.9 0.640 31 pos
190 5 139 80 35 160 31.6 0.361 25 pos
191 3 111 62 0 0 22.6 0.142 21 neg
192 9 123 70 44 94 33.1 0.374 40 neg
193 7 159 66 0 0 30.4 0.383 36 pos
194 11 135 0 0 0 52.3 0.578 40 pos
195 8 85 55 20 0 24.4 0.136 42 neg
196 5 158 84 41 210 39.4 0.395 29 pos
197 1 105 58 0 0 24.3 0.187 21 neg
198 3 107 62 13 48 22.9 0.678 23 pos
199 4 109 64 44 99 34.8 0.905 26 pos
200 4 148 60 27 318 30.9 0.150 29 pos
201 0 113 80 16 0 31.0 0.874 21 neg
202 1 138 82 0 0 40.1 0.236 28 neg
203 0 108 68 20 0 27.3 0.787 32 neg
204 2 99 70 16 44 20.4 0.235 27 neg
205 6 103 72 32 190 37.7 0.324 55 neg
206 5 111 72 28 0 23.9 0.407 27 neg
207 8 196 76 29 280 37.5 0.605 57 pos
208 5 162 104 0 0 37.7 0.151 52 pos
209 1 96 64 27 87 33.2 0.289 21 neg
210 7 184 84 33 0 35.5 0.355 41 pos
211 2 81 60 22 0 27.7 0.290 25 neg
212 0 147 85 54 0 42.8 0.375 24 neg
213 7 179 95 31 0 34.2 0.164 60 neg
214 0 140 65 26 130 42.6 0.431 24 pos
215 9 112 82 32 175 34.2 0.260 36 pos
216 12 151 70 40 271 41.8 0.742 38 pos
217 5 109 62 41 129 35.8 0.514 25 pos
218 6 125 68 30 120 30.0 0.464 32 neg
219 5 85 74 22 0 29.0 1.224 32 pos
220 5 112 66 0 0 37.8 0.261 41 pos
221 0 177 60 29 478 34.6 1.072 21 pos
222 2 158 90 0 0 31.6 0.805 66 pos
223 7 119 0 0 0 25.2 0.209 37 neg
224 7 142 60 33 190 28.8 0.687 61 neg
225 1 100 66 15 56 23.6 0.666 26 neg
226 1 87 78 27 32 34.6 0.101 22 neg
227 0 101 76 0 0 35.7 0.198 26 neg
228 3 162 52 38 0 37.2 0.652 24 pos
229 4 197 70 39 744 36.7 2.329 31 neg
230 0 117 80 31 53 45.2 0.089 24 neg
231 4 142 86 0 0 44.0 0.645 22 pos
232 6 134 80 37 370 46.2 0.238 46 pos
233 1 79 80 25 37 25.4 0.583 22 neg
234 4 122 68 0 0 35.0 0.394 29 neg
235 3 74 68 28 45 29.7 0.293 23 neg
236 4 171 72 0 0 43.6 0.479 26 pos
237 7 181 84 21 192 35.9 0.586 51 pos
238 0 179 90 27 0 44.1 0.686 23 pos
239 9 164 84 21 0 30.8 0.831 32 pos
240 0 104 76 0 0 18.4 0.582 27 neg
241 1 91 64 24 0 29.2 0.192 21 neg
242 4 91 70 32 88 33.1 0.446 22 neg
243 3 139 54 0 0 25.6 0.402 22 pos
244 6 119 50 22 176 27.1 1.318 33 pos
245 2 146 76 35 194 38.2 0.329 29 neg
246 9 184 85 15 0 30.0 1.213 49 pos
247 10 122 68 0 0 31.2 0.258 41 neg
248 0 165 90 33 680 52.3 0.427 23 neg
249 9 124 70 33 402 35.4 0.282 34 neg
250 1 111 86 19 0 30.1 0.143 23 neg
251 9 106 52 0 0 31.2 0.380 42 neg
252 2 129 84 0 0 28.0 0.284 27 neg
253 2 90 80 14 55 24.4 0.249 24 neg
254 0 86 68 32 0 35.8 0.238 25 neg
255 12 92 62 7 258 27.6 0.926 44 pos
256 1 113 64 35 0 33.6 0.543 21 pos
257 3 111 56 39 0 30.1 0.557 30 neg
258 2 114 68 22 0 28.7 0.092 25 neg
259 1 193 50 16 375 25.9 0.655 24 neg
260 11 155 76 28 150 33.3 1.353 51 pos
261 3 191 68 15 130 30.9 0.299 34 neg
262 3 141 0 0 0 30.0 0.761 27 pos
263 4 95 70 32 0 32.1 0.612 24 neg
264 3 142 80 15 0 32.4 0.200 63 neg
265 4 123 62 0 0 32.0 0.226 35 pos
266 5 96 74 18 67 33.6 0.997 43 neg
267 0 138 0 0 0 36.3 0.933 25 pos
268 2 128 64 42 0 40.0 1.101 24 neg
269 0 102 52 0 0 25.1 0.078 21 neg
270 2 146 0 0 0 27.5 0.240 28 pos
271 10 101 86 37 0 45.6 1.136 38 pos
272 2 108 62 32 56 25.2 0.128 21 neg
273 3 122 78 0 0 23.0 0.254 40 neg
274 1 71 78 50 45 33.2 0.422 21 neg
275 13 106 70 0 0 34.2 0.251 52 neg
276 2 100 70 52 57 40.5 0.677 25 neg
277 7 106 60 24 0 26.5 0.296 29 pos
278 0 104 64 23 116 27.8 0.454 23 neg
279 5 114 74 0 0 24.9 0.744 57 neg
280 2 108 62 10 278 25.3 0.881 22 neg
281 0 146 70 0 0 37.9 0.334 28 pos
282 10 129 76 28 122 35.9 0.280 39 neg
283 7 133 88 15 155 32.4 0.262 37 neg
284 7 161 86 0 0 30.4 0.165 47 pos
285 2 108 80 0 0 27.0 0.259 52 pos
286 7 136 74 26 135 26.0 0.647 51 neg
287 5 155 84 44 545 38.7 0.619 34 neg
288 1 119 86 39 220 45.6 0.808 29 pos
289 4 96 56 17 49 20.8 0.340 26 neg
290 5 108 72 43 75 36.1 0.263 33 neg
291 0 78 88 29 40 36.9 0.434 21 neg
292 0 107 62 30 74 36.6 0.757 25 pos
293 2 128 78 37 182 43.3 1.224 31 pos
294 1 128 48 45 194 40.5 0.613 24 pos
295 0 161 50 0 0 21.9 0.254 65 neg
296 6 151 62 31 120 35.5 0.692 28 neg
297 2 146 70 38 360 28.0 0.337 29 pos
298 0 126 84 29 215 30.7 0.520 24 neg
299 14 100 78 25 184 36.6 0.412 46 pos
300 8 112 72 0 0 23.6 0.840 58 neg
301 0 167 0 0 0 32.3 0.839 30 pos
302 2 144 58 33 135 31.6 0.422 25 pos
303 5 77 82 41 42 35.8 0.156 35 neg
304 5 115 98 0 0 52.9 0.209 28 pos
305 3 150 76 0 0 21.0 0.207 37 neg
306 2 120 76 37 105 39.7 0.215 29 neg
307 10 161 68 23 132 25.5 0.326 47 pos
308 0 137 68 14 148 24.8 0.143 21 neg
309 0 128 68 19 180 30.5 1.391 25 pos
310 2 124 68 28 205 32.9 0.875 30 pos
311 6 80 66 30 0 26.2 0.313 41 neg
312 0 106 70 37 148 39.4 0.605 22 neg
313 2 155 74 17 96 26.6 0.433 27 pos
314 3 113 50 10 85 29.5 0.626 25 neg
315 7 109 80 31 0 35.9 1.127 43 pos
316 2 112 68 22 94 34.1 0.315 26 neg
317 3 99 80 11 64 19.3 0.284 30 neg
318 3 182 74 0 0 30.5 0.345 29 pos
319 3 115 66 39 140 38.1 0.150 28 neg
320 6 194 78 0 0 23.5 0.129 59 pos
321 4 129 60 12 231 27.5 0.527 31 neg
322 3 112 74 30 0 31.6 0.197 25 pos
323 0 124 70 20 0 27.4 0.254 36 pos
324 13 152 90 33 29 26.8 0.731 43 pos
325 2 112 75 32 0 35.7 0.148 21 neg
326 1 157 72 21 168 25.6 0.123 24 neg
327 1 122 64 32 156 35.1 0.692 30 pos
328 10 179 70 0 0 35.1 0.200 37 neg
329 2 102 86 36 120 45.5 0.127 23 pos
330 6 105 70 32 68 30.8 0.122 37 neg
331 8 118 72 19 0 23.1 1.476 46 neg
332 2 87 58 16 52 32.7 0.166 25 neg
333 1 180 0 0 0 43.3 0.282 41 pos
334 12 106 80 0 0 23.6 0.137 44 neg
335 1 95 60 18 58 23.9 0.260 22 neg
336 0 165 76 43 255 47.9 0.259 26 neg
337 0 117 0 0 0 33.8 0.932 44 neg
338 5 115 76 0 0 31.2 0.343 44 pos
339 9 152 78 34 171 34.2 0.893 33 pos
340 7 178 84 0 0 39.9 0.331 41 pos
341 1 130 70 13 105 25.9 0.472 22 neg
342 1 95 74 21 73 25.9 0.673 36 neg
343 1 0 68 35 0 32.0 0.389 22 neg
344 5 122 86 0 0 34.7 0.290 33 neg
345 8 95 72 0 0 36.8 0.485 57 neg
346 8 126 88 36 108 38.5 0.349 49 neg
347 1 139 46 19 83 28.7 0.654 22 neg
348 3 116 0 0 0 23.5 0.187 23 neg
349 3 99 62 19 74 21.8 0.279 26 neg
350 5 0 80 32 0 41.0 0.346 37 pos
351 4 92 80 0 0 42.2 0.237 29 neg
352 4 137 84 0 0 31.2 0.252 30 neg
353 3 61 82 28 0 34.4 0.243 46 neg
354 1 90 62 12 43 27.2 0.580 24 neg
355 3 90 78 0 0 42.7 0.559 21 neg
356 9 165 88 0 0 30.4 0.302 49 pos
357 1 125 50 40 167 33.3 0.962 28 pos
358 13 129 0 30 0 39.9 0.569 44 pos
359 12 88 74 40 54 35.3 0.378 48 neg
360 1 196 76 36 249 36.5 0.875 29 pos
361 5 189 64 33 325 31.2 0.583 29 pos
362 5 158 70 0 0 29.8 0.207 63 neg
363 5 103 108 37 0 39.2 0.305 65 neg
364 4 146 78 0 0 38.5 0.520 67 pos
365 4 147 74 25 293 34.9 0.385 30 neg
366 5 99 54 28 83 34.0 0.499 30 neg
367 6 124 72 0 0 27.6 0.368 29 pos
368 0 101 64 17 0 21.0 0.252 21 neg
369 3 81 86 16 66 27.5 0.306 22 neg
370 1 133 102 28 140 32.8 0.234 45 pos
371 3 173 82 48 465 38.4 2.137 25 pos
372 0 118 64 23 89 0.0 1.731 21 neg
373 0 84 64 22 66 35.8 0.545 21 neg
374 2 105 58 40 94 34.9 0.225 25 neg
375 2 122 52 43 158 36.2 0.816 28 neg
376 12 140 82 43 325 39.2 0.528 58 pos
377 0 98 82 15 84 25.2 0.299 22 neg
378 1 87 60 37 75 37.2 0.509 22 neg
379 4 156 75 0 0 48.3 0.238 32 pos
380 0 93 100 39 72 43.4 1.021 35 neg
381 1 107 72 30 82 30.8 0.821 24 neg
382 0 105 68 22 0 20.0 0.236 22 neg
383 1 109 60 8 182 25.4 0.947 21 neg
384 1 90 62 18 59 25.1 1.268 25 neg
385 1 125 70 24 110 24.3 0.221 25 neg
386 1 119 54 13 50 22.3 0.205 24 neg
387 5 116 74 29 0 32.3 0.660 35 pos
388 8 105 100 36 0 43.3 0.239 45 pos
389 5 144 82 26 285 32.0 0.452 58 pos
390 3 100 68 23 81 31.6 0.949 28 neg
391 1 100 66 29 196 32.0 0.444 42 neg
392 5 166 76 0 0 45.7 0.340 27 pos
393 1 131 64 14 415 23.7 0.389 21 neg
394 4 116 72 12 87 22.1 0.463 37 neg
395 4 158 78 0 0 32.9 0.803 31 pos
396 2 127 58 24 275 27.7 1.600 25 neg
397 3 96 56 34 115 24.7 0.944 39 neg
398 0 131 66 40 0 34.3 0.196 22 pos
399 3 82 70 0 0 21.1 0.389 25 neg
400 3 193 70 31 0 34.9 0.241 25 pos
401 4 95 64 0 0 32.0 0.161 31 pos
402 6 137 61 0 0 24.2 0.151 55 neg
403 5 136 84 41 88 35.0 0.286 35 pos
404 9 72 78 25 0 31.6 0.280 38 neg
405 5 168 64 0 0 32.9 0.135 41 pos
406 2 123 48 32 165 42.1 0.520 26 neg
407 4 115 72 0 0 28.9 0.376 46 pos
408 0 101 62 0 0 21.9 0.336 25 neg
409 8 197 74 0 0 25.9 1.191 39 pos
410 1 172 68 49 579 42.4 0.702 28 pos
411 6 102 90 39 0 35.7 0.674 28 neg
412 1 112 72 30 176 34.4 0.528 25 neg
413 1 143 84 23 310 42.4 1.076 22 neg
414 1 143 74 22 61 26.2 0.256 21 neg
415 0 138 60 35 167 34.6 0.534 21 pos
416 3 173 84 33 474 35.7 0.258 22 pos
417 1 97 68 21 0 27.2 1.095 22 neg
418 4 144 82 32 0 38.5 0.554 37 pos
419 1 83 68 0 0 18.2 0.624 27 neg
420 3 129 64 29 115 26.4 0.219 28 pos
421 1 119 88 41 170 45.3 0.507 26 neg
422 2 94 68 18 76 26.0 0.561 21 neg
423 0 102 64 46 78 40.6 0.496 21 neg
424 2 115 64 22 0 30.8 0.421 21 neg
425 8 151 78 32 210 42.9 0.516 36 pos
426 4 184 78 39 277 37.0 0.264 31 pos
427 0 94 0 0 0 0.0 0.256 25 neg
428 1 181 64 30 180 34.1 0.328 38 pos
429 0 135 94 46 145 40.6 0.284 26 neg
430 1 95 82 25 180 35.0 0.233 43 pos
431 2 99 0 0 0 22.2 0.108 23 neg
432 3 89 74 16 85 30.4 0.551 38 neg
433 1 80 74 11 60 30.0 0.527 22 neg
434 2 139 75 0 0 25.6 0.167 29 neg
435 1 90 68 8 0 24.5 1.138 36 neg
436 0 141 0 0 0 42.4 0.205 29 pos
437 12 140 85 33 0 37.4 0.244 41 neg
438 5 147 75 0 0 29.9 0.434 28 neg
439 1 97 70 15 0 18.2 0.147 21 neg
440 6 107 88 0 0 36.8 0.727 31 neg
441 0 189 104 25 0 34.3 0.435 41 pos
442 2 83 66 23 50 32.2 0.497 22 neg
443 4 117 64 27 120 33.2 0.230 24 neg
444 8 108 70 0 0 30.5 0.955 33 pos
445 4 117 62 12 0 29.7 0.380 30 pos
446 0 180 78 63 14 59.4 2.420 25 pos
447 1 100 72 12 70 25.3 0.658 28 neg
448 0 95 80 45 92 36.5 0.330 26 neg
449 0 104 64 37 64 33.6 0.510 22 pos
450 0 120 74 18 63 30.5 0.285 26 neg
451 1 82 64 13 95 21.2 0.415 23 neg
452 2 134 70 0 0 28.9 0.542 23 pos
453 0 91 68 32 210 39.9 0.381 25 neg
454 2 119 0 0 0 19.6 0.832 72 neg
455 2 100 54 28 105 37.8 0.498 24 neg
456 14 175 62 30 0 33.6 0.212 38 pos
457 1 135 54 0 0 26.7 0.687 62 neg
458 5 86 68 28 71 30.2 0.364 24 neg
459 10 148 84 48 237 37.6 1.001 51 pos
460 9 134 74 33 60 25.9 0.460 81 neg
461 9 120 72 22 56 20.8 0.733 48 neg
462 1 71 62 0 0 21.8 0.416 26 neg
463 8 74 70 40 49 35.3 0.705 39 neg
464 5 88 78 30 0 27.6 0.258 37 neg
465 10 115 98 0 0 24.0 1.022 34 neg
466 0 124 56 13 105 21.8 0.452 21 neg
467 0 74 52 10 36 27.8 0.269 22 neg
468 0 97 64 36 100 36.8 0.600 25 neg
469 8 120 0 0 0 30.0 0.183 38 pos
470 6 154 78 41 140 46.1 0.571 27 neg
471 1 144 82 40 0 41.3 0.607 28 neg
472 0 137 70 38 0 33.2 0.170 22 neg
473 0 119 66 27 0 38.8 0.259 22 neg
474 7 136 90 0 0 29.9 0.210 50 neg
475 4 114 64 0 0 28.9 0.126 24 neg
476 0 137 84 27 0 27.3 0.231 59 neg
477 2 105 80 45 191 33.7 0.711 29 pos
478 7 114 76 17 110 23.8 0.466 31 neg
479 8 126 74 38 75 25.9 0.162 39 neg
480 4 132 86 31 0 28.0 0.419 63 neg
481 3 158 70 30 328 35.5 0.344 35 pos
482 0 123 88 37 0 35.2 0.197 29 neg
483 4 85 58 22 49 27.8 0.306 28 neg
484 0 84 82 31 125 38.2 0.233 23 neg
485 0 145 0 0 0 44.2 0.630 31 pos
486 0 135 68 42 250 42.3 0.365 24 pos
487 1 139 62 41 480 40.7 0.536 21 neg
488 0 173 78 32 265 46.5 1.159 58 neg
489 4 99 72 17 0 25.6 0.294 28 neg
490 8 194 80 0 0 26.1 0.551 67 neg
491 2 83 65 28 66 36.8 0.629 24 neg
492 2 89 90 30 0 33.5 0.292 42 neg
493 4 99 68 38 0 32.8 0.145 33 neg
494 4 125 70 18 122 28.9 1.144 45 pos
495 3 80 0 0 0 0.0 0.174 22 neg
496 6 166 74 0 0 26.6 0.304 66 neg
497 5 110 68 0 0 26.0 0.292 30 neg
498 2 81 72 15 76 30.1 0.547 25 neg
499 7 195 70 33 145 25.1 0.163 55 pos
500 6 154 74 32 193 29.3 0.839 39 neg
501 2 117 90 19 71 25.2 0.313 21 neg
502 3 84 72 32 0 37.2 0.267 28 neg
503 6 0 68 41 0 39.0 0.727 41 pos
504 7 94 64 25 79 33.3 0.738 41 neg
505 3 96 78 39 0 37.3 0.238 40 neg
506 10 75 82 0 0 33.3 0.263 38 neg
507 0 180 90 26 90 36.5 0.314 35 pos
508 1 130 60 23 170 28.6 0.692 21 neg
509 2 84 50 23 76 30.4 0.968 21 neg
510 8 120 78 0 0 25.0 0.409 64 neg
511 12 84 72 31 0 29.7 0.297 46 pos
512 0 139 62 17 210 22.1 0.207 21 neg
513 9 91 68 0 0 24.2 0.200 58 neg
514 2 91 62 0 0 27.3 0.525 22 neg
515 3 99 54 19 86 25.6 0.154 24 neg
516 3 163 70 18 105 31.6 0.268 28 pos
517 9 145 88 34 165 30.3 0.771 53 pos
518 7 125 86 0 0 37.6 0.304 51 neg
519 13 76 60 0 0 32.8 0.180 41 neg
520 6 129 90 7 326 19.6 0.582 60 neg
521 2 68 70 32 66 25.0 0.187 25 neg
522 3 124 80 33 130 33.2 0.305 26 neg
523 6 114 0 0 0 0.0 0.189 26 neg
524 9 130 70 0 0 34.2 0.652 45 pos
525 3 125 58 0 0 31.6 0.151 24 neg
526 3 87 60 18 0 21.8 0.444 21 neg
527 1 97 64 19 82 18.2 0.299 21 neg
528 3 116 74 15 105 26.3 0.107 24 neg
529 0 117 66 31 188 30.8 0.493 22 neg
530 0 111 65 0 0 24.6 0.660 31 neg
531 2 122 60 18 106 29.8 0.717 22 neg
532 0 107 76 0 0 45.3 0.686 24 neg
533 1 86 66 52 65 41.3 0.917 29 neg
534 6 91 0 0 0 29.8 0.501 31 neg
535 1 77 56 30 56 33.3 1.251 24 neg
536 4 132 0 0 0 32.9 0.302 23 pos
537 0 105 90 0 0 29.6 0.197 46 neg
538 0 57 60 0 0 21.7 0.735 67 neg
539 0 127 80 37 210 36.3 0.804 23 neg
540 3 129 92 49 155 36.4 0.968 32 pos
541 8 100 74 40 215 39.4 0.661 43 pos
542 3 128 72 25 190 32.4 0.549 27 pos
543 10 90 85 32 0 34.9 0.825 56 pos
544 4 84 90 23 56 39.5 0.159 25 neg
545 1 88 78 29 76 32.0 0.365 29 neg
546 8 186 90 35 225 34.5 0.423 37 pos
547 5 187 76 27 207 43.6 1.034 53 pos
548 4 131 68 21 166 33.1 0.160 28 neg
549 1 164 82 43 67 32.8 0.341 50 neg
550 4 189 110 31 0 28.5 0.680 37 neg
551 1 116 70 28 0 27.4 0.204 21 neg
552 3 84 68 30 106 31.9 0.591 25 neg
553 6 114 88 0 0 27.8 0.247 66 neg
554 1 88 62 24 44 29.9 0.422 23 neg
555 1 84 64 23 115 36.9 0.471 28 neg
556 7 124 70 33 215 25.5 0.161 37 neg
557 1 97 70 40 0 38.1 0.218 30 neg
558 8 110 76 0 0 27.8 0.237 58 neg
559 11 103 68 40 0 46.2 0.126 42 neg
560 11 85 74 0 0 30.1 0.300 35 neg
561 6 125 76 0 0 33.8 0.121 54 pos
562 0 198 66 32 274 41.3 0.502 28 pos
563 1 87 68 34 77 37.6 0.401 24 neg
564 6 99 60 19 54 26.9 0.497 32 neg
565 0 91 80 0 0 32.4 0.601 27 neg
566 2 95 54 14 88 26.1 0.748 22 neg
567 1 99 72 30 18 38.6 0.412 21 neg
568 6 92 62 32 126 32.0 0.085 46 neg
569 4 154 72 29 126 31.3 0.338 37 neg
570 0 121 66 30 165 34.3 0.203 33 pos
571 3 78 70 0 0 32.5 0.270 39 neg
572 2 130 96 0 0 22.6 0.268 21 neg
573 3 111 58 31 44 29.5 0.430 22 neg
574 2 98 60 17 120 34.7 0.198 22 neg
575 1 143 86 30 330 30.1 0.892 23 neg
576 1 119 44 47 63 35.5 0.280 25 neg
577 6 108 44 20 130 24.0 0.813 35 neg
578 2 118 80 0 0 42.9 0.693 21 pos
579 10 133 68 0 0 27.0 0.245 36 neg
580 2 197 70 99 0 34.7 0.575 62 pos
581 0 151 90 46 0 42.1 0.371 21 pos
582 6 109 60 27 0 25.0 0.206 27 neg
583 12 121 78 17 0 26.5 0.259 62 neg
584 8 100 76 0 0 38.7 0.190 42 neg
585 8 124 76 24 600 28.7 0.687 52 pos
586 1 93 56 11 0 22.5 0.417 22 neg
587 8 143 66 0 0 34.9 0.129 41 pos
588 6 103 66 0 0 24.3 0.249 29 neg
589 3 176 86 27 156 33.3 1.154 52 pos
590 0 73 0 0 0 21.1 0.342 25 neg
591 11 111 84 40 0 46.8 0.925 45 pos
592 2 112 78 50 140 39.4 0.175 24 neg
593 3 132 80 0 0 34.4 0.402 44 pos
594 2 82 52 22 115 28.5 1.699 25 neg
595 6 123 72 45 230 33.6 0.733 34 neg
596 0 188 82 14 185 32.0 0.682 22 pos
597 0 67 76 0 0 45.3 0.194 46 neg
598 1 89 24 19 25 27.8 0.559 21 neg
599 1 173 74 0 0 36.8 0.088 38 pos
600 1 109 38 18 120 23.1 0.407 26 neg
601 1 108 88 19 0 27.1 0.400 24 neg
602 6 96 0 0 0 23.7 0.190 28 neg
603 1 124 74 36 0 27.8 0.100 30 neg
604 7 150 78 29 126 35.2 0.692 54 pos
605 4 183 0 0 0 28.4 0.212 36 pos
606 1 124 60 32 0 35.8 0.514 21 neg
607 1 181 78 42 293 40.0 1.258 22 pos
608 1 92 62 25 41 19.5 0.482 25 neg
609 0 152 82 39 272 41.5 0.270 27 neg
610 1 111 62 13 182 24.0 0.138 23 neg
611 3 106 54 21 158 30.9 0.292 24 neg
612 3 174 58 22 194 32.9 0.593 36 pos
613 7 168 88 42 321 38.2 0.787 40 pos
614 6 105 80 28 0 32.5 0.878 26 neg
615 11 138 74 26 144 36.1 0.557 50 pos
616 3 106 72 0 0 25.8 0.207 27 neg
617 6 117 96 0 0 28.7 0.157 30 neg
618 2 68 62 13 15 20.1 0.257 23 neg
619 9 112 82 24 0 28.2 1.282 50 pos
620 0 119 0 0 0 32.4 0.141 24 pos
621 2 112 86 42 160 38.4 0.246 28 neg
622 2 92 76 20 0 24.2 1.698 28 neg
623 6 183 94 0 0 40.8 1.461 45 neg
624 0 94 70 27 115 43.5 0.347 21 neg
625 2 108 64 0 0 30.8 0.158 21 neg
626 4 90 88 47 54 37.7 0.362 29 neg
627 0 125 68 0 0 24.7 0.206 21 neg
628 0 132 78 0 0 32.4 0.393 21 neg
629 5 128 80 0 0 34.6 0.144 45 neg
630 4 94 65 22 0 24.7 0.148 21 neg
631 7 114 64 0 0 27.4 0.732 34 pos
632 0 102 78 40 90 34.5 0.238 24 neg
633 2 111 60 0 0 26.2 0.343 23 neg
634 1 128 82 17 183 27.5 0.115 22 neg
635 10 92 62 0 0 25.9 0.167 31 neg
636 13 104 72 0 0 31.2 0.465 38 pos
637 5 104 74 0 0 28.8 0.153 48 neg
638 2 94 76 18 66 31.6 0.649 23 neg
639 7 97 76 32 91 40.9 0.871 32 pos
640 1 100 74 12 46 19.5 0.149 28 neg
641 0 102 86 17 105 29.3 0.695 27 neg
642 4 128 70 0 0 34.3 0.303 24 neg
643 6 147 80 0 0 29.5 0.178 50 pos
644 4 90 0 0 0 28.0 0.610 31 neg
645 3 103 72 30 152 27.6 0.730 27 neg
646 2 157 74 35 440 39.4 0.134 30 neg
647 1 167 74 17 144 23.4 0.447 33 pos
648 0 179 50 36 159 37.8 0.455 22 pos
649 11 136 84 35 130 28.3 0.260 42 pos
650 0 107 60 25 0 26.4 0.133 23 neg
651 1 91 54 25 100 25.2 0.234 23 neg
652 1 117 60 23 106 33.8 0.466 27 neg
653 5 123 74 40 77 34.1 0.269 28 neg
654 2 120 54 0 0 26.8 0.455 27 neg
655 1 106 70 28 135 34.2 0.142 22 neg
656 2 155 52 27 540 38.7 0.240 25 pos
657 2 101 58 35 90 21.8 0.155 22 neg
658 1 120 80 48 200 38.9 1.162 41 neg
659 11 127 106 0 0 39.0 0.190 51 neg
660 3 80 82 31 70 34.2 1.292 27 pos
661 10 162 84 0 0 27.7 0.182 54 neg
662 1 199 76 43 0 42.9 1.394 22 pos
663 8 167 106 46 231 37.6 0.165 43 pos
664 9 145 80 46 130 37.9 0.637 40 pos
665 6 115 60 39 0 33.7 0.245 40 pos
666 1 112 80 45 132 34.8 0.217 24 neg
667 4 145 82 18 0 32.5 0.235 70 pos
668 10 111 70 27 0 27.5 0.141 40 pos
669 6 98 58 33 190 34.0 0.430 43 neg
670 9 154 78 30 100 30.9 0.164 45 neg
671 6 165 68 26 168 33.6 0.631 49 neg
672 1 99 58 10 0 25.4 0.551 21 neg
673 10 68 106 23 49 35.5 0.285 47 neg
674 3 123 100 35 240 57.3 0.880 22 neg
675 8 91 82 0 0 35.6 0.587 68 neg
676 6 195 70 0 0 30.9 0.328 31 pos
677 9 156 86 0 0 24.8 0.230 53 pos
678 0 93 60 0 0 35.3 0.263 25 neg
679 3 121 52 0 0 36.0 0.127 25 pos
680 2 101 58 17 265 24.2 0.614 23 neg
681 2 56 56 28 45 24.2 0.332 22 neg
682 0 162 76 36 0 49.6 0.364 26 pos
683 0 95 64 39 105 44.6 0.366 22 neg
684 4 125 80 0 0 32.3 0.536 27 pos
685 5 136 82 0 0 0.0 0.640 69 neg
686 2 129 74 26 205 33.2 0.591 25 neg
687 3 130 64 0 0 23.1 0.314 22 neg
688 1 107 50 19 0 28.3 0.181 29 neg
689 1 140 74 26 180 24.1 0.828 23 neg
690 1 144 82 46 180 46.1 0.335 46 pos
691 8 107 80 0 0 24.6 0.856 34 neg
692 13 158 114 0 0 42.3 0.257 44 pos
693 2 121 70 32 95 39.1 0.886 23 neg
694 7 129 68 49 125 38.5 0.439 43 pos
695 2 90 60 0 0 23.5 0.191 25 neg
696 7 142 90 24 480 30.4 0.128 43 pos
697 3 169 74 19 125 29.9 0.268 31 pos
698 0 99 0 0 0 25.0 0.253 22 neg
699 4 127 88 11 155 34.5 0.598 28 neg
700 4 118 70 0 0 44.5 0.904 26 neg
701 2 122 76 27 200 35.9 0.483 26 neg
702 6 125 78 31 0 27.6 0.565 49 pos
703 1 168 88 29 0 35.0 0.905 52 pos
704 2 129 0 0 0 38.5 0.304 41 neg
705 4 110 76 20 100 28.4 0.118 27 neg
706 6 80 80 36 0 39.8 0.177 28 neg
707 10 115 0 0 0 0.0 0.261 30 pos
708 2 127 46 21 335 34.4 0.176 22 neg
709 9 164 78 0 0 32.8 0.148 45 pos
710 2 93 64 32 160 38.0 0.674 23 pos
711 3 158 64 13 387 31.2 0.295 24 neg
712 5 126 78 27 22 29.6 0.439 40 neg
713 10 129 62 36 0 41.2 0.441 38 pos
714 0 134 58 20 291 26.4 0.352 21 neg
715 3 102 74 0 0 29.5 0.121 32 neg
716 7 187 50 33 392 33.9 0.826 34 pos
717 3 173 78 39 185 33.8 0.970 31 pos
718 10 94 72 18 0 23.1 0.595 56 neg
719 1 108 60 46 178 35.5 0.415 24 neg
720 5 97 76 27 0 35.6 0.378 52 pos
721 4 83 86 19 0 29.3 0.317 34 neg
722 1 114 66 36 200 38.1 0.289 21 neg
723 1 149 68 29 127 29.3 0.349 42 pos
724 5 117 86 30 105 39.1 0.251 42 neg
725 1 111 94 0 0 32.8 0.265 45 neg
726 4 112 78 40 0 39.4 0.236 38 neg
727 1 116 78 29 180 36.1 0.496 25 neg
728 0 141 84 26 0 32.4 0.433 22 neg
729 2 175 88 0 0 22.9 0.326 22 neg
730 2 92 52 0 0 30.1 0.141 22 neg
731 3 130 78 23 79 28.4 0.323 34 pos
732 8 120 86 0 0 28.4 0.259 22 pos
733 2 174 88 37 120 44.5 0.646 24 pos
734 2 106 56 27 165 29.0 0.426 22 neg
735 2 105 75 0 0 23.3 0.560 53 neg
736 4 95 60 32 0 35.4 0.284 28 neg
737 0 126 86 27 120 27.4 0.515 21 neg
738 8 65 72 23 0 32.0 0.600 42 neg
739 2 99 60 17 160 36.6 0.453 21 neg
740 1 102 74 0 0 39.5 0.293 42 pos
741 11 120 80 37 150 42.3 0.785 48 pos
742 3 102 44 20 94 30.8 0.400 26 neg
743 1 109 58 18 116 28.5 0.219 22 neg
744 9 140 94 0 0 32.7 0.734 45 pos
745 13 153 88 37 140 40.6 1.174 39 neg
746 12 100 84 33 105 30.0 0.488 46 neg
747 1 147 94 41 0 49.3 0.358 27 pos
748 1 81 74 41 57 46.3 1.096 32 neg
749 3 187 70 22 200 36.4 0.408 36 pos
750 6 162 62 0 0 24.3 0.178 50 pos
751 4 136 70 0 0 31.2 1.182 22 pos
752 1 121 78 39 74 39.0 0.261 28 neg
753 3 108 62 24 0 26.0 0.223 25 neg
754 0 181 88 44 510 43.3 0.222 26 pos
755 8 154 78 32 0 32.4 0.443 45 pos
756 1 128 88 39 110 36.5 1.057 37 pos
757 7 137 90 41 0 32.0 0.391 39 neg
758 0 123 72 0 0 36.3 0.258 52 pos
759 1 106 76 0 0 37.5 0.197 26 neg
760 6 190 92 0 0 35.5 0.278 66 pos
761 2 88 58 26 16 28.4 0.766 22 neg
762 9 170 74 31 0 44.0 0.403 43 pos
763 9 89 62 0 0 22.5 0.142 33 neg
764 10 101 76 48 180 32.9 0.171 63 neg
765 2 122 70 27 0 36.8 0.340 27 neg
766 5 121 72 23 112 26.2 0.245 30 neg
767 1 126 60 0 0 30.1 0.349 47 pos
768 1 93 70 31 0 30.4 0.315 23 negA quick exploration reveals that there are more zeros in the data than expected (especially since a BMI or tricep skin fold thickness of 0 is impossible), implying that missing values are recorded as zeros. See for instance the histogram of the tricep skin fold thickness, which has a number of 0 entries that are set apart from the other entries.
ggplot(diabetes_orig) +
geom_histogram(aes(x = triceps))`stat_bin()` using `bins = 30`. Pick better value with `binwidth`.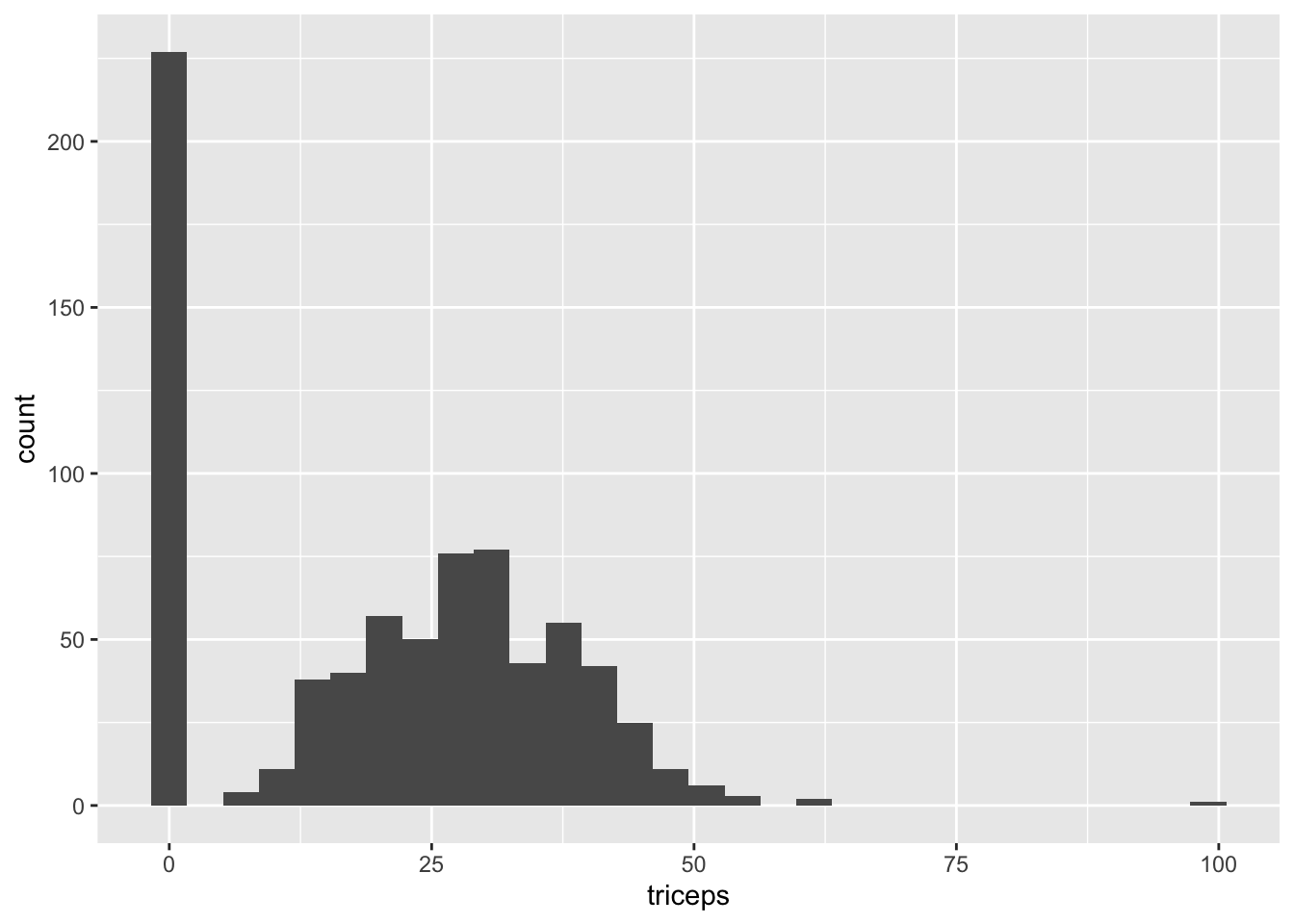
This phenomena can also seen in the glucose, pressure, insulin and mass variables. Thus, we convert the 0 entries in all variables (other than “pregnant”) to NA. To do that, we use the mutate_at() function (which will soon be superseded by mutate() with across()) to specify which variables we want to apply our mutating function to, and we use the if_else() function to specify what to replace the value with if the condition is true or false.
diabetes_clean <- diabetes_orig %>%
mutate_at(vars(triceps, glucose, pressure, insulin, mass),
function(.var) {
if_else(condition = (.var == 0), # if true (i.e. the entry is 0)
true = as.numeric(NA), # replace the value with NA
false = .var # otherwise leave it as it is
)
})Our data is ready. Hopefully you’ve replenished your cup of tea (or coffee if you’re into that for some reason). Let’s start making some tidy models!
Split into train/test
First, let’s split our dataset into training and testing data. The training data will be used to fit our model and tune its parameters, where the testing data will be used to evaluate our final model’s performance.
This split can be done automatically using the inital_split() function (from rsample) which creates a special “split” object.
set.seed(234589)
# split the data into trainng (75%) and testing (25%)
diabetes_split <- initial_split(diabetes_clean,
prop = 3/4)
diabetes_split<Training/Testing/Total>
<576/192/768>The printed output of diabetes_split, our split object, tells us how many observations we have in the training set, the testing set, and overall: <train/test/total>.
The training and testing sets can be extracted from the “split” object using the training() and testing() functions. Although, we won’t actually use these objects in the pipeline (we will be using the diabetes_split object itself).
# extract training and testing sets
diabetes_train <- training(diabetes_split)
diabetes_test <- testing(diabetes_split)At some point we’re going to want to do some parameter tuning, and to do that we’re going to want to use cross-validation. So we can create a cross-validated version of the training set in preparation for that moment using vfold_cv().
# create CV object from training data
diabetes_cv <- vfold_cv(diabetes_train)Define a recipe
Recipes allow you to specify the role of each variable as an outcome or predictor variable (using a “formula”), and any pre-processing steps you want to conduct (such as normalization, imputation, PCA, etc).
Creating a recipe has two parts (layered on top of one another using pipes %>%):
Specify the formula (
recipe()): specify the outcome variable and predictor variablesSpecify pre-processing steps (
step_zzz()): define the pre-processing steps, such as imputation, creating dummy variables, scaling, and more
For instance, we can define the following recipe
# define the recipe
diabetes_recipe <-
# which consists of the formula (outcome ~ predictors)
recipe(diabetes ~ pregnant + glucose + pressure + triceps +
insulin + mass + pedigree + age,
data = diabetes_clean) %>%
# and some pre-processing steps
step_normalize(all_numeric()) %>%
step_impute_knn(all_predictors())If you’ve ever seen formulas before (e.g. using the lm() function in R), you might have noticed that we could have written our formula much more efficiently using the formula short-hand where . represents all of the variables in the data: outcome ~ . will fit a model that predicts the outcome using all other columns.
The full list of pre-processing steps available can be found here. In the recipe steps above we used the functions all_numeric() and all_predictors() as arguments to the pre-processing steps. These are called “role selections”, and they specify that we want to apply the step to “all numeric” variables or “all predictor variables”. The list of all potential role selectors can be found by typing ?selections into your console.
Note that we used the original diabetes_clean data object (we set recipe(..., data = diabetes_clean)), rather than the diabetes_train object or the diabetes_split object. It turns out we could have used any of these. All recipes takes from the data object at this point is the names and roles of the outcome and predictor variables. We will apply this recipe to specific datasets later. This means that for large data sets, the head of the data could be used to pass the recipe a smaller data set to save time and memory.
Indeed, if we print a summary of the diabetes_recipe object, it just shows us how many predictor variables we’ve specified and the steps we’ve specified (but it doesn’t actually implement them yet!).
diabetes_recipe── Recipe ──────────────────────────────────────────────────────────────────────── Inputs Number of variables by roleoutcome: 1
predictor: 8── Operations • Centering and scaling for: all_numeric()• K-nearest neighbor imputation for: all_predictors()If you want to extract the pre-processed dataset itself, you can first prep() the recipe for a specific dataset and juice() the prepped recipe to extract the pre-processed data. It turns out that extracting the pre-processed data isn’t actually necessary for the pipeline, since this will be done under the hood when the model is fit, but sometimes it’s useful anyway.
diabetes_train_preprocessed <- diabetes_recipe %>%
# apply the recipe to the training data
prep(diabetes_train) %>%
# extract the pre-processed training dataset
juice()
diabetes_train_preprocessed# A tibble: 576 × 9
pregnant glucose pressure triceps insulin mass pedigree age diabetes
<dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <fct>
1 1.23 -0.390 0.262 0.892 -0.317 -0.656 0.502 -0.201 pos
2 0.0447 1.09 -0.0616 -0.0348 -0.219 -0.168 -0.400 0.301 neg
3 1.82 1.91 -0.224 0.595 0.146 0.379 -0.813 0.301 neg
4 -1.14 -0.0948 -0.710 -0.591 -0.522 -0.311 3.76 -1.04 neg
5 -0.548 0.233 0.424 0.706 0.239 1.56 2.25 -0.201 pos
6 0.637 -0.0620 -1.84 -0.683 0.190 -0.771 2.53 -0.0335 pos
7 -0.844 -1.64 -2.01 -1.05 -0.629 -1.73 -0.445 -0.953 neg
8 -0.548 -0.423 -1.68 -0.313 -0.735 0.00505 -0.460 -0.953 neg
9 -1.14 0.463 -0.386 1.17 0.796 1.41 -0.320 -0.786 pos
10 1.53 1.41 0.424 0.150 0.877 0.0482 -0.968 0.969 pos
# … with 566 more rowsI wrote a much longer post on recipes if you’d like to check out more details. However, note that the preparation and bake steps described in that post are no longer necessary in the tidymodels pipeline, since they’re now implemented under the hood by the later model fitting functions in this pipeline.
Specify the model
So far we’ve split our data into training/testing, and we’ve specified our pre-processing steps using a recipe. The next thing we want to specify is our model (using the parsnip package).
Parsnip offers a unified interface for the massive variety of models that exist in R. This means that you only have to learn one way of specifying a model, and you can use this specification and have it generate a linear model, a random forest model, a support vector machine model, and more with a single line of code.
There are a few primary components that you need to provide for the model specification
The model type: what kind of model you want to fit, set using a different function depending on the model, such as
rand_forest()for random forest,logistic_reg()for logistic regression,svm_poly()for a polynomial SVM model etc. The full list of models available via parsnip can be found here.The arguments: the model parameter values (now consistently named across different models), set using
set_args().The engine: the underlying package the model should come from (e.g. “ranger” for the ranger implementation of Random Forest), set using
set_engine().The mode: the type of prediction - since several packages can do both classification (binary/categorical prediction) and regression (continuous prediction), set using
set_mode().
For instance, if we want to fit a random forest model as implemented by the ranger package for the purpose of classification and we want to tune the mtry parameter (the number of randomly selected variables to be considered at each split in the trees), then we would define the following model specification:
rf_model <-
# specify that the model is a random forest
rand_forest() %>%
# specify that the `mtry` parameter needs to be tuned
set_args(mtry = tune()) %>%
# select the engine/package that underlies the model
set_engine("ranger", importance = "impurity") %>%
# choose either the continuous regression or binary classification mode
set_mode("classification") If you want to be able to examine the variable importance of your final model later, you will need to set importance argument when setting the engine. For ranger, the importance options are "impurity" or "permutation".
As another example, the following code would instead specify a logistic regression model from the glm package.
lr_model <-
# specify that the model is a random forest
logistic_reg() %>%
# select the engine/package that underlies the model
set_engine("glm") %>%
# choose either the continuous regression or binary classification mode
set_mode("classification") Note that this code doesn’t actually fit the model. Like the recipe, it just outlines a description of the model. Moreover, setting a parameter to tune() means that it will be tuned later in the tune stage of the pipeline (i.e. the value of the parameter that yields the best performance will be chosen). You could also just specify a particular value of the parameter if you don’t want to tune it e.g. using set_args(mtry = 4).
Another thing to note is that nothing about this model specification is specific to the diabetes dataset.
Put it all together in a workflow
We’re now ready to put the model and recipes together into a workflow. You initiate a workflow using workflow() (from the workflows package) and then you can add a recipe and add a model to it.
# set the workflow
rf_workflow <- workflow() %>%
# add the recipe
add_recipe(diabetes_recipe) %>%
# add the model
add_model(rf_model)Note that we still haven’t yet implemented the pre-processing steps in the recipe nor have we fit the model. We’ve just written the framework. It is only when we tune the parameters or fit the model that the recipe and model frameworks are actually implemented.
Tune the parameters
Since we had a parameter that we designated to be tuned (mtry), we need to tune it (i.e. choose the value that leads to the best performance) before fitting our model. If you don’t have any parameters to tune, you can skip this step.
Note that we will do our tuning using the cross-validation object (diabetes_cv). To do this, we specify the range of mtry values we want to try, and then we add a tuning layer to our workflow using tune_grid() (from the tune package). Note that we focus on two metrics: accuracy and roc_auc (from the yardstick package).
# specify which values eant to try
rf_grid <- expand.grid(mtry = c(3, 4, 5))
# extract results
rf_tune_results <- rf_workflow %>%
tune_grid(resamples = diabetes_cv, #CV object
grid = rf_grid, # grid of values to try
metrics = metric_set(accuracy, roc_auc) # metrics we care about
)You can tune multiple parameters at once by providing multiple parameters to the expand.grid() function, e.g. expand.grid(mtry = c(3, 4, 5), trees = c(100, 500)).
It’s always a good idea to explore the results of the cross-validation. collect_metrics() is a really handy function that can be used in a variety of circumstances to extract any metrics that have been calculated within the object it’s being used on. In this case, the metrics come from the cross-validation performance across the different values of the parameters.
# print results
rf_tune_results %>%
collect_metrics()# A tibble: 6 × 7
mtry .metric .estimator mean n std_err .config
<dbl> <chr> <chr> <dbl> <int> <dbl> <chr>
1 3 accuracy binary 0.762 10 0.0108 Preprocessor1_Model1
2 3 roc_auc binary 0.840 10 0.0116 Preprocessor1_Model1
3 4 accuracy binary 0.767 10 0.0130 Preprocessor1_Model2
4 4 roc_auc binary 0.840 10 0.0128 Preprocessor1_Model2
5 5 accuracy binary 0.766 10 0.0108 Preprocessor1_Model3
6 5 roc_auc binary 0.837 10 0.0118 Preprocessor1_Model3Across both accuracy and AUC, mtry = 4 yields the best performance (just).
Finalize the workflow
We want to add a layer to our workflow that corresponds to the tuned parameter, i.e. sets mtry to be the value that yielded the best results. If you didn’t tune any parameters, you can skip this step.
We can extract the best value for the accuracy metric by applying the select_best() function to the tune object.
param_final <- rf_tune_results %>%
select_best(metric = "accuracy")
param_final# A tibble: 1 × 2
mtry .config
<dbl> <chr>
1 4 Preprocessor1_Model2Then we can add this parameter to the workflow using the finalize_workflow() function.
rf_workflow <- rf_workflow %>%
finalize_workflow(param_final)Evaluate the model on the test set
Now we’ve defined our recipe, our model, and tuned the model’s parameters, we’re ready to actually fit the final model. Since all of this information is contained within the workflow object, we will apply the last_fit() function to our workflow and our train/test split object. This will automatically train the model specified by the workflow using the training data, and produce evaluations based on the test set.
rf_fit <- rf_workflow %>%
# fit on the training set and evaluate on test set
last_fit(diabetes_split)Note that the fit object that is created is a data-frame-like object; specifically, it is a tibble with list columns.
rf_fit# Resampling results
# Manual resampling
# A tibble: 1 × 6
splits id .metrics .notes .predictions .workflow
<list> <chr> <list> <list> <list> <list>
1 <split [576/192]> train/test split <tibble> <tibble> <tibble> <workflow>This is a really nice feature of tidymodels (and is what makes it work so nicely with the tidyverse) since you can do all of your tidyverse operations to the model object. While truly taking advantage of this flexibility requires proficiency with purrr, if you don’t want to deal with purrr and list-columns, there are functions that can extract the relevant information from the fit object that remove the need for purrr as we will see below.
Since we supplied the train/test object when we fit the workflow, the metrics are evaluated on the test set. Now when we use the collect_metrics() function (recall we used this when tuning our parameters), it extracts the performance of the final model (since rf_fit now consists of a single final model) applied to the test set.
test_performance <- rf_fit %>% collect_metrics()
test_performance# A tibble: 2 × 4
.metric .estimator .estimate .config
<chr> <chr> <dbl> <chr>
1 accuracy binary 0.792 Preprocessor1_Model1
2 roc_auc binary 0.853 Preprocessor1_Model1Overall the performance is very good, with an accuracy of 0.74 and an AUC of 0.82.
You can also extract the test set predictions themselves using the collect_predictions() function. Note that there are 192 rows in the predictions object below which matches the number of test set observations (just to give you some evidence that these are based on the test set rather than the training set).
# generate predictions from the test set
test_predictions <- rf_fit %>% collect_predictions()
test_predictions# A tibble: 192 × 7
id .pred_neg .pred_pos .row .pred_class diabetes .config
<chr> <dbl> <dbl> <int> <fct> <fct> <chr>
1 train/test split 0.365 0.635 3 pos pos Preprocessor…
2 train/test split 0.213 0.787 9 pos pos Preprocessor…
3 train/test split 0.765 0.235 11 neg neg Preprocessor…
4 train/test split 0.511 0.489 13 neg neg Preprocessor…
5 train/test split 0.594 0.406 18 neg pos Preprocessor…
6 train/test split 0.356 0.644 26 pos pos Preprocessor…
7 train/test split 0.209 0.791 32 pos pos Preprocessor…
8 train/test split 0.751 0.249 50 neg neg Preprocessor…
9 train/test split 0.961 0.0385 53 neg neg Preprocessor…
10 train/test split 0.189 0.811 55 pos neg Preprocessor…
# … with 182 more rowsSince this is just a normal data frame/tibble object, we can generate summaries and plots such as a confusion matrix.
# generate a confusion matrix
test_predictions %>%
conf_mat(truth = diabetes, estimate = .pred_class) Truth
Prediction neg pos
neg 107 17
pos 23 45We could also plot distributions of the predicted probability distributions for each class.
test_predictions %>%
ggplot() +
geom_density(aes(x = .pred_pos, fill = diabetes),
alpha = 0.5)
If you’re familiar with purrr, you could use purrr functions to extract the predictions column using pull(). The following code does almost the same thing as collect_predictions(). You could similarly have done this with the .metrics column.
test_predictions <- rf_fit %>% pull(.predictions)
test_predictions[[1]]
# A tibble: 192 × 6
.pred_neg .pred_pos .row .pred_class diabetes .config
<dbl> <dbl> <int> <fct> <fct> <chr>
1 0.365 0.635 3 pos pos Preprocessor1_Model1
2 0.213 0.787 9 pos pos Preprocessor1_Model1
3 0.765 0.235 11 neg neg Preprocessor1_Model1
4 0.511 0.489 13 neg neg Preprocessor1_Model1
5 0.594 0.406 18 neg pos Preprocessor1_Model1
6 0.356 0.644 26 pos pos Preprocessor1_Model1
7 0.209 0.791 32 pos pos Preprocessor1_Model1
8 0.751 0.249 50 neg neg Preprocessor1_Model1
9 0.961 0.0385 53 neg neg Preprocessor1_Model1
10 0.189 0.811 55 pos neg Preprocessor1_Model1
# … with 182 more rowsFitting and using your final model
The previous section evaluated the model trained on the training data using the testing data. But once you’ve determined your final model, you often want to train it on your full dataset and then use it to predict the response for new data.
If you want to use your model to predict the response for new observations, you need to use the fit() function on your workflow and the dataset that you want to fit the final model on (e.g. the complete training + testing dataset).
final_model <- fit(rf_workflow, diabetes_clean)The final_model object contains a few things including the ranger object trained with the parameters established through the workflow contained in rf_workflow based on the data in diabetes_clean (the combined training and testing data).
final_model══ Workflow [trained] ══════════════════════════════════════════════════════════
Preprocessor: Recipe
Model: rand_forest()
── Preprocessor ────────────────────────────────────────────────────────────────
2 Recipe Steps
• step_normalize()
• step_impute_knn()
── Model ───────────────────────────────────────────────────────────────────────
Ranger result
Call:
ranger::ranger(x = maybe_data_frame(x), y = y, mtry = min_cols(~4, x), importance = ~"impurity", num.threads = 1, verbose = FALSE, seed = sample.int(10^5, 1), probability = TRUE)
Type: Probability estimation
Number of trees: 500
Sample size: 768
Number of independent variables: 8
Mtry: 4
Target node size: 10
Variable importance mode: impurity
Splitrule: gini
OOB prediction error (Brier s.): 0.1583874 If we wanted to predict the diabetes status of a new woman, we could use the normal predict() function.
For instance, below we define the data for a new woman.
new_woman <- tribble(~pregnant, ~glucose, ~pressure, ~triceps, ~insulin, ~mass, ~pedigree, ~age,
2, 95, 70, 31, 102, 28.2, 0.67, 47)
new_woman# A tibble: 1 × 8
pregnant glucose pressure triceps insulin mass pedigree age
<dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
1 2 95 70 31 102 28.2 0.67 47The predicted diabetes status of this new woman is “negative”.
predict(final_model, new_data = new_woman)# A tibble: 1 × 1
.pred_class
<fct>
1 neg Variable importance
If you want to extract the variable importance scores from your model, as far as I can tell, for now you need to extract the model object from the fit() object (which for us is called final_model). The function that extracts the model is pull_workflow_fit() and then you need to grab the fit object that the output contains.
ranger_obj <- pull_workflow_fit(final_model)$fitWarning: `pull_workflow_fit()` was deprecated in workflows 0.2.3.
ℹ Please use `extract_fit_parsnip()` instead.ranger_objRanger result
Call:
ranger::ranger(x = maybe_data_frame(x), y = y, mtry = min_cols(~4, x), importance = ~"impurity", num.threads = 1, verbose = FALSE, seed = sample.int(10^5, 1), probability = TRUE)
Type: Probability estimation
Number of trees: 500
Sample size: 768
Number of independent variables: 8
Mtry: 4
Target node size: 10
Variable importance mode: impurity
Splitrule: gini
OOB prediction error (Brier s.): 0.1583874 Then you can extract the variable importance from the ranger object itself (variable.importance is a specific object contained within ranger output - this will need to be adapted for the specific object type of other models).
ranger_obj$variable.importancepregnant glucose pressure triceps insulin mass pedigree age
16.40687 79.68408 17.08361 22.10685 52.27195 42.60717 30.12246 33.19040